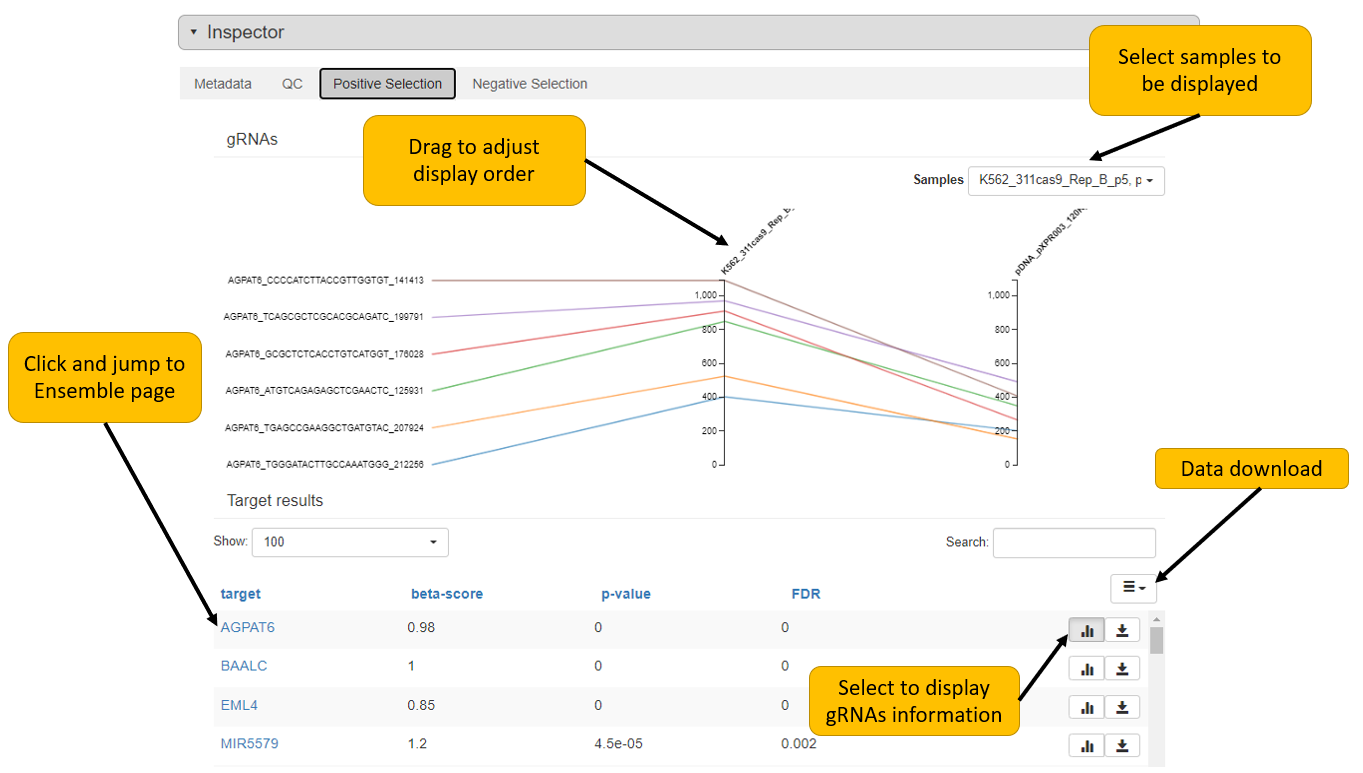
1. Search your interested gene
CRISP-view mainly provides two options to search the database. One is gene searching, the other is biological source, including cell line, tissue, publication (PMID) and treatment condition. User can explore the essentiality of interested gene by gene searching, and view detailed data annotations, quality control report and positively/negatively selected genes of individual sample by clicking selected bar. The waterfall plot is ranked beta-score of searched gene in matched samples. Beta-score is a measurement of gene selections similar to the term of log fold change in differential expression analysis. A positive beta-core means a gene is positively selected in a condition, vice versa. Detailed explanation of beta-score please refer to
MAGeCK-VISPR paper.
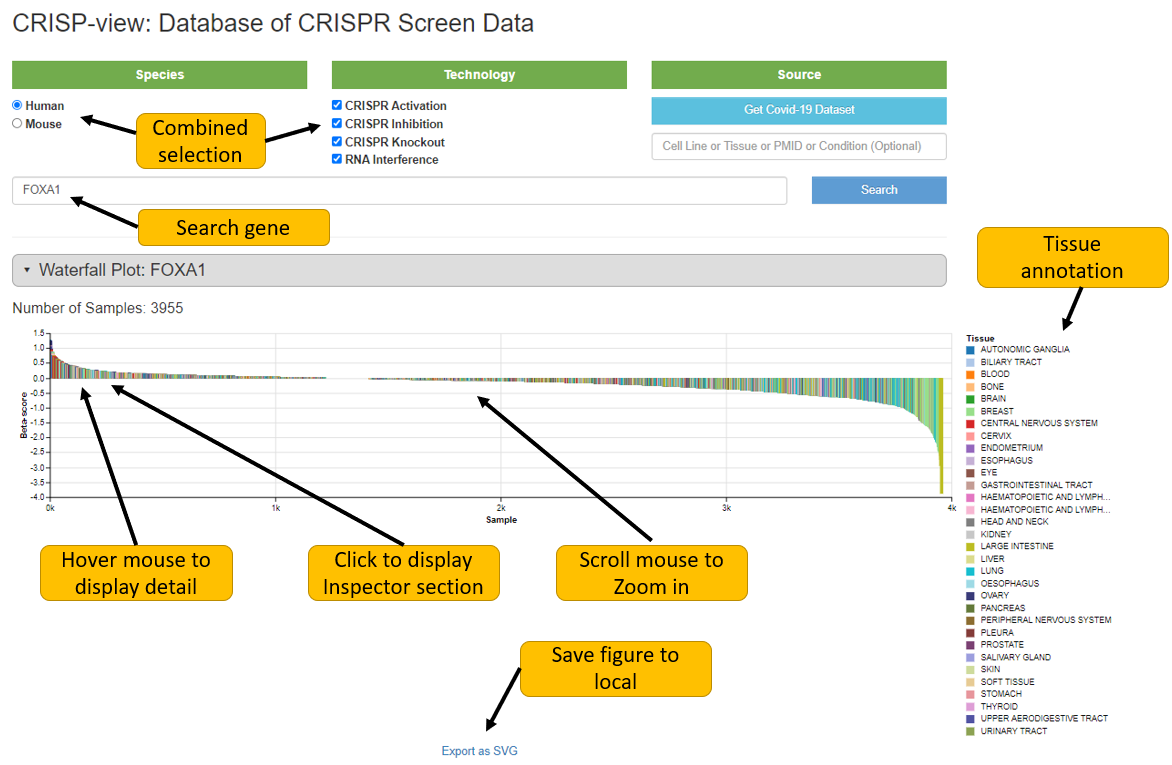
2. Search your interested biological source
User can explore interested cell line, tissue, publication or treatment condition by biological source searching. Matched samples will be listed in a table. User can view detailed data annotations, quality control report, positively/negatively selected genes of individual sample by clicking selected sample. The quality control report is shown here briefly by colored circles. Green circle means passing the quality control test, red circle means failing the quality control test, and grey circle means not applicable for this sample. The meaning of each circles will appear while mouse hovers above the circle.
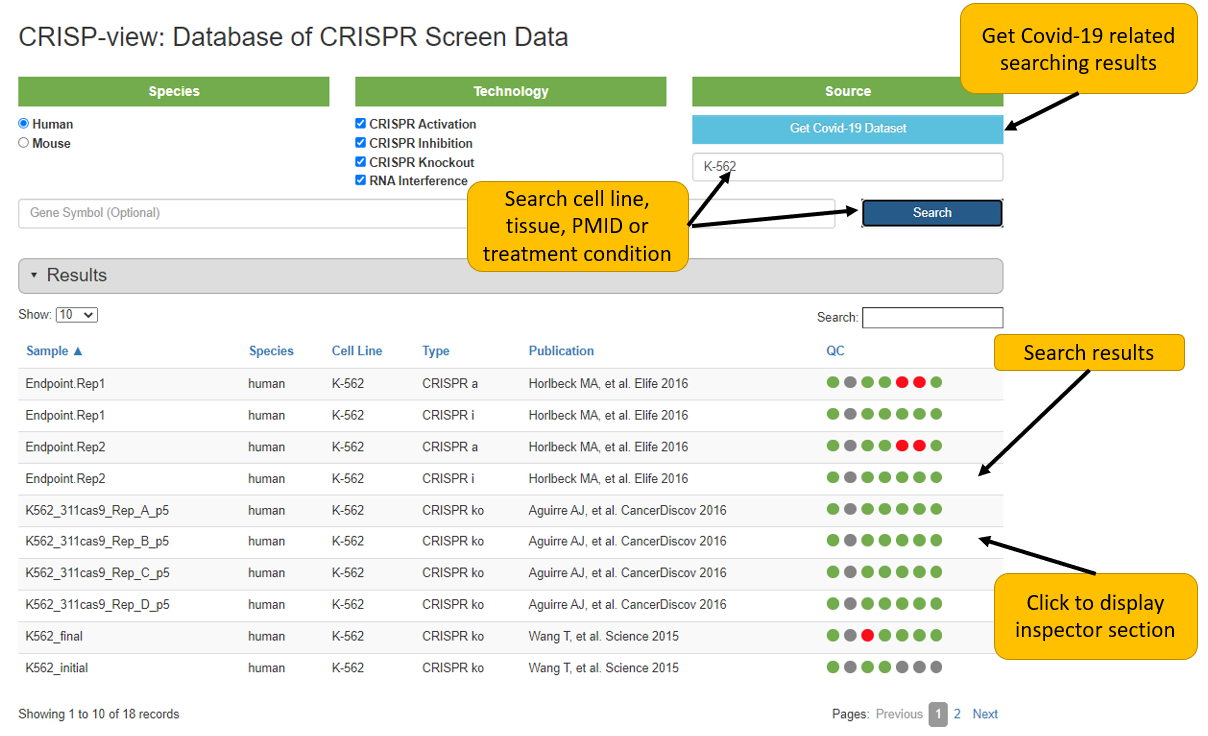
3. Inspect individual dataset
After clicking interested sample, inspector section for that sample will be shown as follows. The inspector section displays the detailed data annotations, quality control report and positively/negatively selected genes of individual sample in separated tabs.
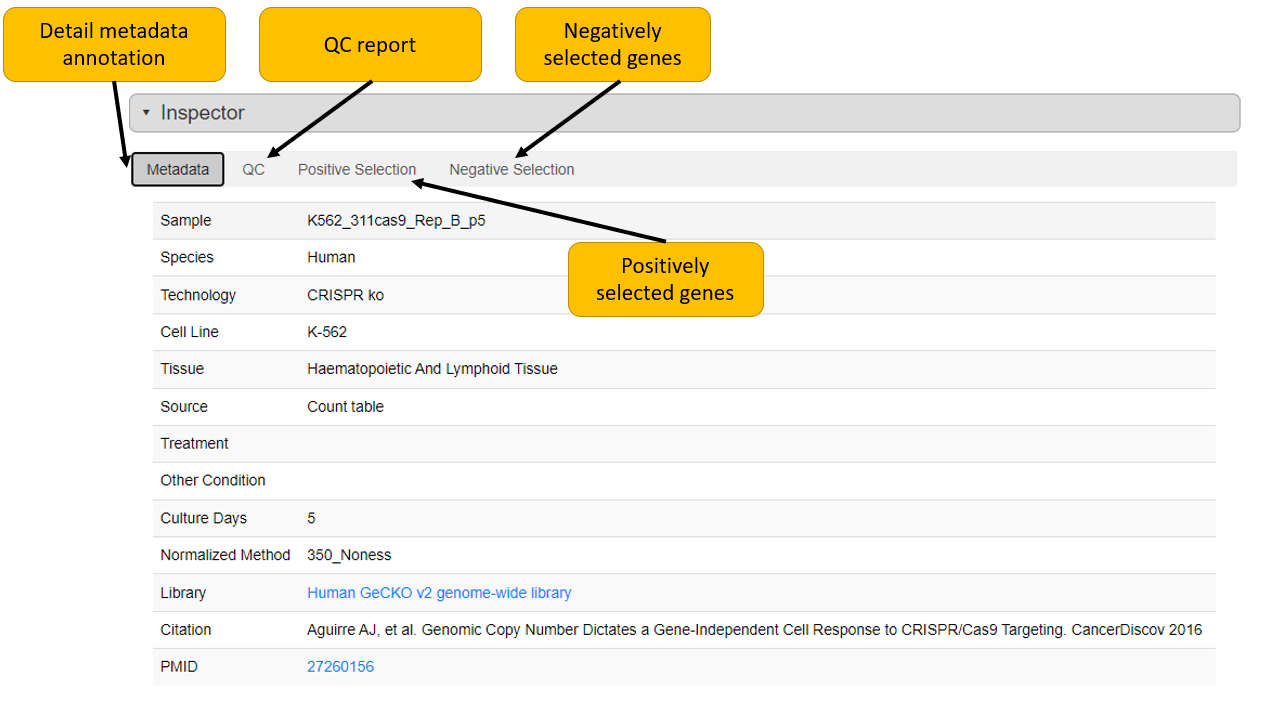
4. Inspect positively/negative selected genes
The positive/negative selection tabs of inspector section show corresponding genes in list in order of beta-score. User can view read count of gRNAs of selected sample and its corresponding initial condition(s) here. User can also download gene selection information as tab-separated text file here.